P3E-EGFPpA: Difference between revisions
Jump to navigation
Jump to search
Hggenusrwki (talk | contribs) No edit summary |
Hggenusrwki (talk | contribs) No edit summary |
||
| Line 8: | Line 8: | ||
== Crude Map == | == Crude Map == | ||
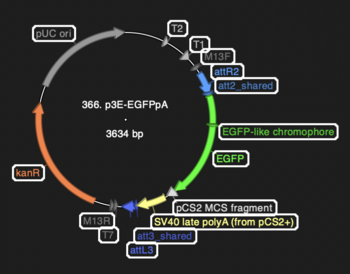
Screenshot from Sequencher showing locations of components: | Screenshot from Sequencher showing locations of components: | ||
:[[File:366 p3E-EGFPpA.PNG|thumb|none|350px|366 p3E-EGFPpA]] | |||
==Download the Sequence == | ==Download the Sequence == | ||
* '''Annotated Sequence'''(Genbank format) and '''Full-Length Sequence with individual component sequences included'''(FASTA file) | * '''Annotated Sequence'''(Genbank format) and '''Full-Length Sequence with individual component sequences included'''(FASTA file) | ||
: [[Media:366.p3E-EGFPpA.zip|Download p3E-EGFPpA]] | : [[Media:366.p3E-EGFPpA.zip|Download p3E-EGFPpA]] | ||
Revision as of 18:20, 29 May 2025
Construction Details
p3E-EGFPpA was made by PCR amplification of EGFP and an SV40 late polyadenylation signal from a pCS2+-based plasmid, using primers to add att sites, followed by a BP reaction.
Note that the EGFP sequence is followed by 60 bp of polylinker sequence from pCS2+ before the stop codon. We have confirmed functionally that this cassette produces fluorescent protein in C-terminal fusion constructs. Sequencing confirms that all bases of the insert are correct.
The EGFP sequence does include a start codon and a good Kozak consensus, so it is possible to use this clone to test promoters cloned into a middle clone.
Crude Map
Screenshot from Sequencher showing locations of components:
Download the Sequence
- Annotated Sequence(Genbank format) and Full-Length Sequence with individual component sequences included(FASTA file)