PCS2FA-transposase: Difference between revisions
Jump to navigation
Jump to search
Hggenusrwki (talk | contribs) Created page with "== Construction Details == This template is used to display DNA sequence information in a standardized format. == Crude Map == photo showing locations of components == Sequence == * Annotated Sequence: (Genbank format) link * Full-Length Sequence with individual component sequences included: (FASTA file) link == Reference == This template is used to display DNA sequence information in a standardized format." |
No edit summary |
||
| (14 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
== Construction Details == | == Construction Details == | ||
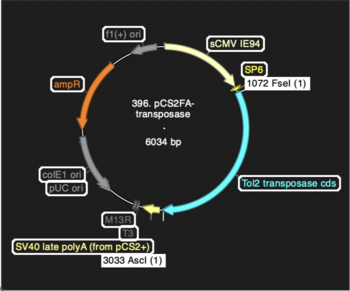
pCS2FA-transposase was made by PCRing the Tol2 transpoase cds from the original Kawakami clone, adding restriction sites, then conventionally subcloned into pCS2FA, a version of pCS2+ modified by adding FseI and AscI sites. Sequencing confirms that all bases of the insert are correct. | |||
The original transposase clone included start and stop codons upstream of the transposase cds, and a polyA sequence (AAAAA) rather than a polyadenylation ''signal''. | |||
== Crude Map == | == Crude Map == | ||
Screenshot from ApE showing locations of components: | |||
:[[File:396 pCS2FA-Tol2transposase.PNG|thumb|none|350px|396 pCS2FA-Tol2transposase]] | |||
== | ==Download the Sequence == | ||
* '''Annotated Sequence'''(Genbank format) and '''Full-Length Sequence with individual component sequences included'''(FASTA file) | |||
: [[Media:396.pCS2FA-transposase.zip|Download pCS2FA-transposase]] | |||
Latest revision as of 20:01, 29 May 2025
Construction Details
pCS2FA-transposase was made by PCRing the Tol2 transpoase cds from the original Kawakami clone, adding restriction sites, then conventionally subcloned into pCS2FA, a version of pCS2+ modified by adding FseI and AscI sites. Sequencing confirms that all bases of the insert are correct.
The original transposase clone included start and stop codons upstream of the transposase cds, and a polyA sequence (AAAAA) rather than a polyadenylation signal.
Crude Map
Screenshot from ApE showing locations of components:
Download the Sequence
- Annotated Sequence(Genbank format) and Full-Length Sequence with individual component sequences included(FASTA file)