P5E-bactin2: Difference between revisions
Hggenusrwki (talk | contribs) No edit summary |
No edit summary |
||
| (9 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
== Construction Details == | == Construction Details == | ||
''bactin2'' genomic sequence was derived from a 5.3 kb clone (gift of Ken Poss), which was derived in turn from a 10 kb upstream construct from Shinichi Higashijima. | |||
p5E-bactin2 was made by PCR of the bactin2 upstream genomic sequence (using primers to add att sites), followed by a BP reaction. Upstream sequence includes 3.8 kb upstream of transcriptional start, a small 5' UTR exon, a 1.4 kb intron, and then 38 bp of 5' UTR, terminating immediately before the native start codon. Sequencing confirms that the ends of the insert are correct. | |||
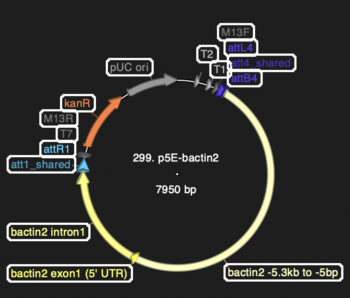
== Crude Map == | |||
Screenshot from ApE showing locations of components: | |||
:[[File:299 p5E-bactin2.PNG|thumb|none|350px|299 p5E-bactin2]] | |||
==Download the Sequence == | ==Download the Sequence == | ||
* Annotated Sequence (Genbank format) and Full-Length Sequence with individual component sequences included (FASTA file) | * '''Annotated Sequence''' (Genbank format) and '''Full-Length Sequence with individual component sequences included''' (FASTA file) | ||
: [[Media: | : [[Media:299.p5E-bactin2.zip|Download p5E-''bactin2'']] | ||
== Reference == | == Reference == | ||
Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. (1997) Dev Biol.15;192:289-99. "High-frequency generation of transgenic zebrafish which | |||
reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin." | |||
Latest revision as of 04:01, 3 June 2025
Construction Details
bactin2 genomic sequence was derived from a 5.3 kb clone (gift of Ken Poss), which was derived in turn from a 10 kb upstream construct from Shinichi Higashijima.
p5E-bactin2 was made by PCR of the bactin2 upstream genomic sequence (using primers to add att sites), followed by a BP reaction. Upstream sequence includes 3.8 kb upstream of transcriptional start, a small 5' UTR exon, a 1.4 kb intron, and then 38 bp of 5' UTR, terminating immediately before the native start codon. Sequencing confirms that the ends of the insert are correct.
Crude Map
Screenshot from ApE showing locations of components:
Download the Sequence
- Annotated Sequence (Genbank format) and Full-Length Sequence with individual component sequences included (FASTA file)
Reference
Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. (1997) Dev Biol.15;192:289-99. "High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin."