PME-MCS: Difference between revisions
Hggenusrwki (talk | contribs) Created page with "== Construction Details == This template is used to display DNA sequence information in a standardized format. == Crude Map == photo showing locations of components == Sequence == * Annotated Sequence: (Genbank format) link * Full-Length Sequence with individual component sequences included: (FASTA file) link == Reference == This template is used to display DNA sequence information in a standardized format." |
No edit summary |
||
| (One intermediate revision by one other user not shown) | |||
| Line 1: | Line 1: | ||
== Construction Details == | == Construction Details == | ||
pME-MCS was made by PCR of the pBSII SK+ multiple cloning site, from M13F to M13R primers, followed by a BP reaction with pDONR221. Sequencing confirms that all bases of the insert are correct. | |||
== | The following sites from the Bluescript MCS remain unique in pME-MCS: | ||
<pre> | |||
''KpnI-XhoI-SalI-Bsp106-HindIII-EcoRI-PstI-SmaI-BamHI-SpeI-XbaI-NotI-XmaIII-SacII-BstXI-SacI'' | |||
</pre> | |||
== Sequencing Primers == | |||
The insert contains the entire M13F sequence, and the 3' end of the M13R sequence. Therefore M13F will bind both in the vector backbone and in the insert, and M13R will bind perfectly to the vector backbone and weakly in the insert. There are also T7 sites in both the insert and the vector backbone. | |||
The T3 site at one end of the insert should be unique and work for sequencing. | |||
Clearly we did not think carefully about sequencing primers when designing this clone. Sorry. | |||
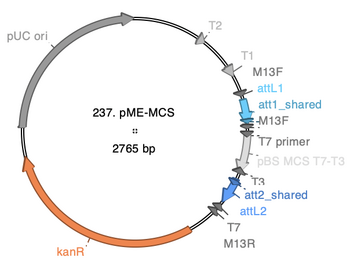
== Crude Map == | |||
Screenshot from ApE showing locations of components: | |||
:[[File:237 pME-MCScrop.PNG|thumb|none|350px|237 pME-MCS]] | |||
== | ==Download the Sequence == | ||
* '''Annotated Sequence'''(Genbank format) and '''Full-Length Sequence with individual component sequences included'''(FASTA file) | |||
: [[Media:237.pME-MCS.zip|Download pME-MCS]] | |||
Latest revision as of 20:40, 29 May 2025
Construction Details
pME-MCS was made by PCR of the pBSII SK+ multiple cloning site, from M13F to M13R primers, followed by a BP reaction with pDONR221. Sequencing confirms that all bases of the insert are correct.
The following sites from the Bluescript MCS remain unique in pME-MCS:
''KpnI-XhoI-SalI-Bsp106-HindIII-EcoRI-PstI-SmaI-BamHI-SpeI-XbaI-NotI-XmaIII-SacII-BstXI-SacI''
Sequencing Primers
The insert contains the entire M13F sequence, and the 3' end of the M13R sequence. Therefore M13F will bind both in the vector backbone and in the insert, and M13R will bind perfectly to the vector backbone and weakly in the insert. There are also T7 sites in both the insert and the vector backbone.
The T3 site at one end of the insert should be unique and work for sequencing.
Clearly we did not think carefully about sequencing primers when designing this clone. Sorry.
Crude Map
Screenshot from ApE showing locations of components:
Download the Sequence
- Annotated Sequence(Genbank format) and Full-Length Sequence with individual component sequences included(FASTA file)