P5E-MCS: Difference between revisions
Jump to navigation
Jump to search
Hggenusrwki (talk | contribs) No edit summary |
No edit summary |
||
| (One intermediate revision by one other user not shown) | |||
| Line 9: | Line 9: | ||
== Crude Map == | == Crude Map == | ||
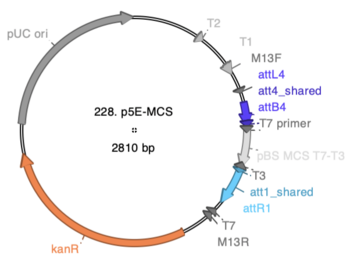
Screenshot from | Screenshot from ApE showing locations of components: | ||
:[[File:228 p5E-MCS.PNG|thumb|none|350px|228 p5E-MCS]] | |||
==Download the Sequence == | ==Download the Sequence == | ||
* '''Annotated Sequence'''(Genbank format) and '''Full-Length Sequence with individual component sequences included'''(FASTA file) | * '''Annotated Sequence'''(Genbank format) and '''Full-Length Sequence with individual component sequences included'''(FASTA file) | ||
: [[Media:228.p5E-MCS.zip|Download p5E-MCS]] | : [[Media:228.p5E-MCS.zip|Download p5E-MCS]] | ||
Latest revision as of 19:55, 29 May 2025
Construction Details
p5E-MCS was made by PCR of the pBSII SK+ multiple cloning site, from T7 to T3 primers, followed by a BP reaction with pDONR P4-P1R. Sequencing confirms that all bases of the insert are correct.
M13F, M13R, and T3 should all work as sequencing primers.
Unfortunately, the backbone of P4-P1R contains recognition sites for several potentially useful restriction enzymes. The restriction sites from the Bluescript MCS that remain unique in p5E-MCS are:
Asp718-KpnI-DraII-XhoI-SalI-Bsp106-HindIII-SmaI-BamHI-SpeI-SacII-BstXI
Crude Map
Screenshot from ApE showing locations of components:
Download the Sequence
- Annotated Sequence(Genbank format) and Full-Length Sequence with individual component sequences included(FASTA file)